Going with the Flow
Chemistry, physics researchers join forces to unravel mystery behind cell signaling
Cells are the fundamental building blocks of life. Humans are composed of trillions of cells, which are responsible for everything from regulating our growth and immune responses to creating hormones, extracting nutrients from food, and providing the structure for our bodies.
Yet, there are still vast gaps in what researchers know about cells—including their signaling methods. Aurelia Honerkamp-Smith, assistant professor of physics, and Damien Thévenin, associate professor of chemistry, are working to change that with help from a five-year, $1.5 million National Institutes of Health grant to define and predict how cells sense and respond to fluid flow. The pair are also examining the method by which lipids and proteins travel in response to fluid flow.
This stage of the project is focused on endothelial cells, which line blood vessels, transport nutrients and gases, and regulate body processes including blood pressure, bone density, and neural growth. When these cells sense healthy blood flow, they produce nitric oxide, which, in turn, increases circulation and helps the blood more efficiently deliver nutrients and oxygen.
When they don’t feel enough flow, the endothelial cells trigger immune responses including inflammation that, if chronic, can lead to heart disease, obesity, cancers, and a host of other health issues—which is why aerobic exercise is so important. A better understanding of the principles of the fluid mechanics behind cell signaling mechanisms, then, could someday lead to therapies that mitigate the development of cardiovascular diseases and other conditions.
The project, in its second year, is exploring the flow-mediated transport of glypican-1, a protein that exists on the exterior and interior of the cell surface and initiates nitric oxide production in response to flow. Discovering the trigger mechanism, however, has been a challenge.
“There’s something of a paradox in the field of flow sensing,” Honerkamp-Smith says. “Most of the time, we think about cells sensing mechanical force using individual proteins that change their shape. For instance, you might have a protein that’s folded up, and then, if a force pulls on it and partially unfolds it, that’s the signal. Or you might have an ion channel that’s closed, then a force pulls on it and opens it and some ions can go through, and that’s your signal. But, in the case of blood flow sensing, nobody really knows what molecule is doing the sensing. The shear force that blood flow applies to blood vessels is too weak to cause this kind of motion in individual proteins.”
But, she says, “what we propose is that even if forces are very weak, they can move membrane proteins along the surface of the cell. Even a very weak force can push on each protein, like wind on a little sailboat, and that way, you could have proteins move over. When the cell notices all the proteins are to one side, it might say, ‘Ah! There’s a flow outside, pushing in that direction.’ Our ultimate goal is to really determine whether that’s what endothelial cells do to sense blood flow.”
A Need to Measure Speed
One of the first steps was to develop a method to measure protein speed, she says.
“We’ve been exploring, in a model system, the movement of proteins under flow. We have a couple of papers out; one is a proof of principle demonstrating that we can make an artificial membrane approximately the size of a single cell—just a flat membrane on a glass surface. Then, we attach proteins and apply flow to it, and we can measure how fast the proteins move according to the amount of shear force we’re applying to them. It’s a new method we've developed … it’s quite sensitive and pretty reliable,” she says.
But, the team also needs to create a way to tag and track the proteins without changing their size or destroying the cells with which they interact.
“On our side, we develop model proteins of different sizes to test whether they move differently in function of flow. We are also trying to develop a way to tag the glypican-1 protein selectively without modifying its size dramatically,” Thévenin says. “We can label the protein with an antibody, but the antibody is gigantic—about three times the size of glypican-1. Conventionally, we could attach GFP, green fluorescent protein, onto it, but it changes the size, affecting its response to flow. So, we’re trying to develop a way to tag the protein on the surface of living cells, without being toxic to cells; without modifying the size of the protein dramatically; and only labeling that precise protein.”
The goal, Thévenin says, is to have two or three differently sized protein models and then determine if size correlates to how they move and respond to flow.
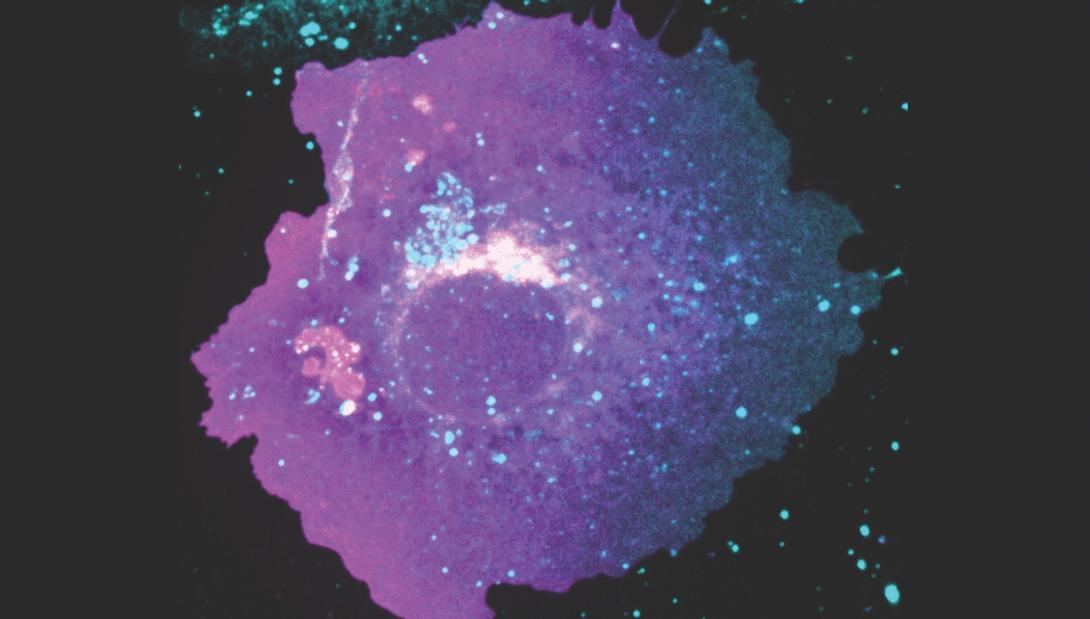
Honerkamp-Smith’s lab then utilizes a confocal fluorescence microscope to observe how the proteins move across the surface of the cells.
“We’re able to collect three-dimensional images of where all these fluorescent proteins are, and we have a microfluidic flow channel, where we can apply a controlled amount of flow and also keep them at the right temperature and in their usual growth medium,” she says. “We’re also developing image analysis code to determine how fast proteins are moving in response to flows.
“What we observed is what we were hoping to observe, which is that larger proteins go faster than smaller ones,” she says. “So, if you have a bigger sail, then you move faster under flow, and we can actually measure the forces that we’re applying to each individual protein. Those are really tiny forces. These are femtonewton-sized forces, which are incredibly small. We’re quite excited about that.”
“I think the big challenge,” Honerkamp-Smith says, “is that in order to see the response, we have to do this with cells that are alive and happy. It’s not as hard to get beautiful images of cells if you’ve killed them and frozen them in place, but we need to do this with living, functioning cells.”
In addition to the size restrictions Thévenin mentioned, the team also needs to find a location for the tag that won’t change the protein’s role.
“We’re trying to get a very specific, very tiny fluorescent label onto these proteins, and then, we can apply flow, watch the protein move and then see if that’s correlated with the initiation of the signal transduction,” she says.
“Everything that has to deal with lipid membranes is extremely difficult to study,” Thévenin says. “One, lipids are hydrophobic, and so the proteins that bind to them and interact with them are also hydrophobic. And two, you need to detect how one single protein, among all the others, moves with very, very tiny forces. It’s like observing the movement of a very small boat in an ocean from very far up.”
Tiny Forces, Large Impact
One of the most significant findings so far, Honerkamp-Smith says, is that “we can get dramatic rearrangement of the whole cell surface with such a tiny force applied to each individual protein. It’s hard to convey how small the forces are … piconewtons are the tiniest forces that people usually deal with, and these forces are 10 times smaller than that! So, we’re really looking at something extremely subtle that has a big effect on the outside of the cell.
“I’m very pleased that we’ve seen protein gradients forming on the outside of living cells that were growing,” she says. “This is something I did expect to find, but it’s exciting to see it in reality. This isn’t published yet, so that’s a very new, in-progress result that we’re working on now.”
Thévenin says the next step will be to look at glypican-1 in living cells and see whether it can be labeled, followed, flowed at different rates, and linked to a difference in cell response.
“There is also part of the grant that is just looking at the lipids, without proteins,” he says. And, initial findings are already eye-opening.
“As we’ve been doing experiments, we see that in addition to moving the proteins, just the lipids by themselves do surprising things in response to flow,” Honerkamp-Smith says. “There are shape changes we’ve observed that we’re still doing experiments on to try to figure out what is happening on a basic level.”
“When you do an experiment, you open a can of worms,” Thévenin says. “You have more questions, more hypotheses to test and even more experiments to do—and that’s where the fun is.”

